Hyper Parameter Optimization (HPO) with Supervisor
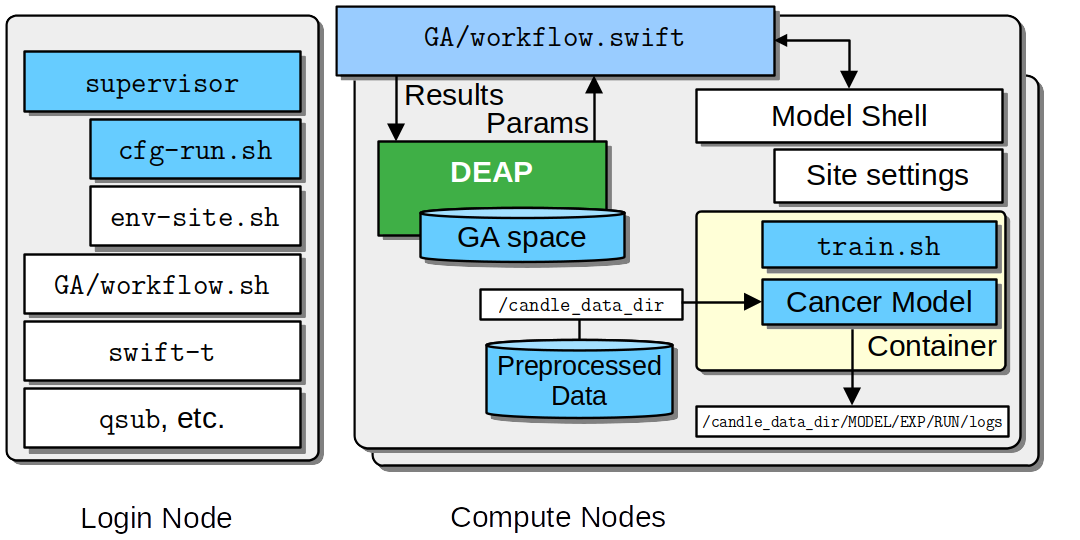
The above figure provides an overview of the newer tool-driven approach using the supervisor command-line tool with cfg scripts and SUPERVISOR_PATH. Login and Compute node internals show the DEAP-driven Genetic Algorithm (GA) workflow for Hyperparameter Optimization (HPO) on any supercomputer. Color Coding: Blue boxes indicate user edits, while white boxes are primarily provided by Supervisor.
HPO Prerequisites
Installing git and singularity
Please refer to the documentation of the tools for install instructions.
Installing supervisor
conda create --name IMPROVE python=3.9.16
conda activate IMPROVE
# change to a directory for supervisor if desired
git clone https://github.com/ECP-CANDLE/Supervisor.git
cd Supervisor && export PATH=$PATH:$(pwd)/bin
git checkout develop
pip install numpy deap
Installing swift-t
You need to be in conda environment to proceed. If you aren’t sure:
# Same conda env than above
# conda create --name IMPROVE python=3.9.16
conda activate IMPROVE
Install swift-t via conda. For detailed instructions please refer to the swift-t documentation:
conda install --yes -c conda-forge -c swift-t swift-t
Setting up a new site
Supervisor/workflows/common/sh/env-local.shFor more complex systems, you will also need to provide scheduler settings in the sched-SITE.sh script.
See the supervisor tool doc for more details about how to configure a site.
Requirements
The following are the requirements as a model curator for others to run HPO on your model.
IMPROVE MODEL (Defined for Containerization)
Your model must be IMPROVE compliant, reading arguments from a ‘.txt’ file and overwriting with command-line arguments. Your model must also be defined in a ‘def’ file for singularity containerization. Default definition files can be found in the IMPROVE Singularity repository. The container should expose the following interface scripts:
preprocess.sh
train.sh
infer.sh
To test your scripts with containerization, it’s recommended you build a container and run the following commands (customized with your arguments):
singularity exec --bind $IMPROVE_DATA_DIR:/IMPROVE_DATA_DIR <path_to_sif_file>.sif preprocess.sh /IMPROVE_DATA_DIR \
--train_split_file <dataset>_split_0_train.txt --val_split_file <dataset>_split_0_val.txt \
--ml_data_outdir /IMPROVE_DATA_DIR/<desired_outdir>
singularity exec --nv --bind $IMPROVE_DATA_DIR:/IMPROVE_DATA_DIR <path_to_sif_file>.sif train.sh <gpu_num> /IMPROVE_DATA_DIR \
--train_ml_data_dir <path> --val_ml_data_dir <dir> --model_outdir <path> --test_ml_data_dir <path>
HYPERPARAMETER SPACE
You will also need to define the hyperparameter space, which will override the arguments in the .txt file. For this reason, any pathing arguments needed for your train script will also need to be defined as ‘constant’ hyperparameters in the space (such as train_ml_data_dir below).
At a high level, the upper and lower describe the bounds of the hyperparameter. Hyperparameters of float, int, ordered, categorical, and constant types are supported, with ordered and categorical hyperparameters supporting float, int, and string types. Log scale exploration is also supported for float and int hyperparameter types.
More specifically, the hyperparameter configuration file has a json format consisting of a list of json dictionaries, each one of which defines a hyperparameter and how it is explored:
Universal Keys
name: the name of the hyperparameter (e.g. _epochs_)
type: determines how the initial population (i.e. the hyperparameter sets) are initialized from the named parameter and how those values are subsequently mutated by the GA. Type is one of constant, int, float, logical, categorical, or ordered.
constant:
each model is initialized with the same specifed value
mutation always returns the same specified value
int:
each model is initialized with an int randomly drawn from the range defined by lower and upper bounds
mutation is peformed by adding the results of a random draw from a gaussian distribution to the current value, where the gaussian distribution’s mu is 0 and its sigma is specified by the sigma entry.
float:
each model is initialized with a float randomly drawn from the range defined by lower and upper bounds
mutation is peformed by adding the results of a random draw from a gaussian distribution to the current value, where the gaussian distribution’s mu is 0 and its sigma is specified by the sigma entry.
logical:
each model is initialized with a random boolean.
mutation flips the logical value
categorical:
each model is initialized with an element chosen at random from the list of elements in values.
mutation chooses an element from the values list at random
ordered:
each model is inititalized with an element chosen at random from the list of elements in values.
given the index of the current value in the list of values, mutation selects the element _n_ number of indices away, where n is the result of a random draw between 1 and sigma and then is negated with a 0.5 probability.
Type Specific Keys
Required
The following keys are required depending on value of the type key.
If the type is constant:
value: the constant value
If the type is int, or float:
lower: the lower bound of the range to draw from
upper: the upper bound of the range to draw from
If the type is categorical:
values: the list of elements to choose from
element_type: the type of the elements to choose from. One of int, float, string, or logical
If the type is ordered:
values: the list of elements to choose from
element_type: the type of the elements to choose from. One of int, float, string, or logical
Optional
The following keys are optional depending on value of the type key.
If the type is constant or float:
use_log_scale: whether to apply mutation on log_10 of the hyperparameter or not
sigma: the sigma value used by the mutation operator. Roughly, it controls the size of mutations (see above).
If the type is ordered:
sigma: the sigma value used by the mutation operator. Roughly, it controls the size of mutations (see above).
Example File
A sample hyperparameter definition file:
[ { "name": "train_ml_data_dir", "type": "constant", "value": "<train_data_dir>" }, { "name": "val_ml_data_dir", "type": "constant", "value": "<val_data_dir>" }, { "name": "model_outdir", "type": "constant", "value": "<desired_outdir>" }, { "name": "learning_rate", "type": "float", "use_log_scale": true, "lower": 0.000001, "upper": 0.0001, "sigma": 0.1 }, { "name": "num_layers", "type": "int", "lower": 1, "upper": 9 }, { "name": "batch_size", "type": "ordered", "element_type": "int", "values": [16, 32, 64, 128, 256, 512], "sigma": 1 }, { "name": "warmup_type", "type": "ordered", "element_type": "string", "values": ["none", "linear", "quadratic", "exponential"] }, { "name": "optimizer", "type": "categorical", "element_type": "string", "values": [ "Adam", "SGD", "RMSprop" ] }, { "name": "epochs", "type": "constant", "value": 150 } ]
Note that any other keys are ignored by the workflow but can be used to add additional information about the hyperparameter. For example, the sample files could contain a comment entry that contains additional information about that hyperparameter and its use.
Requirements for scripts
The following are the requirements to run HPO on your model.
Your model must be IMPROVE compliant, containerized, and packaged in a singularity image. You can identify the image file by the *.sif suffix. Default definition files can be found in the IMPROVE Singularity repository. The container should expose the following interface scripts:
preprocess.sh
train.sh
infer.sh
Steps
Install prerequisites
Preprocess data
Create config files for experiment.
Run HPO with supervisor: supervisor ${location} ${workflow} ${config}
Analysis
Preprocess Data, if needed
This step is only necessary if your data has not already been preprocessed and stored on your filesystem. To preprocess your data, you’ll need to call preprocess.sh in your singularity container with the needed command line arguments. Preprocess using the following command with your arguments:
singularity exec --bind $IMPROVE_DATA_DIR:/IMPROVE_DATA_DIR <path_to_sif_file>.sif preprocess.sh /IMPROVE_DATA_DIR --train_split_file <dataset>_split_0_train.txt \
--val_split_file <dataset>_split_0_val.txt --ml_data_outdir /IMPROVE_DATA_DIR/<desired_outdir>
Create config files
A directory with copy-and-customize config files here at Example Files, along with a README that explains the settings used. Create with the following steps:
mkdir Experiment && cd ExperimentCreate config file cfg-1.sh:
source_cfg -v cfg-my-settings.sh export CANDLE_MODEL_TYPE="SINGULARITY" export MODEL_NAME=${/PATH/TO/SINGULARITY/IMAGE/FILE.sif} export PARAM_SET_FILE=${/PATH/TO/GA/PARAMETER/FILE.json} #e.g hyperparams.json
Create config file cfg-my-settings.sh:
echo SETTINGS # General Settings export PROCS=4 export PPN=4 export WALLTIME=01:00:00 export NUM_ITERATIONS=3 export POPULATION_SIZE=2 # GA Settings (optional) export GA_STRATEGY='mu_plus_lambda' export OFFSPRING_PROPORTION=0.5 export MUT_PROB=0.8 export CX_PROB=0.2 export MUT_INDPB=0.5 export CX_INDPB=0.5 export TOURNSIZE=4 # Add any additional settings needed for your system. General settings and system settings need to be set by the user, while GA settings don't need to be changed. # Default settings for lambda and polaris are given here. # If you have write access to the shared filesystem on your computation system (such as /lambda_stor), # you can save there. If not, make a directory in /tmp or somewhere else you can write. # Lambda Settings # export CANDLE_DATA_DIR=/tmp/<user>/data_dir # Polaris Settings # export QUEUE="debug" # export CANDLE_DATA_DIR=/home/<user>/data_dir
More information on Polaris job submitting (nodes, walltime, queue, etc…) can be found here: https://docs.alcf.anl.gov/polaris/running-jobs/
Create parameter file hyperparams.json:
[ { "name": "train_ml_data_dir", "type": "constant", "value": "<train_data_dir>" }, { "name": "val_ml_data_dir", "type": "constant", "value": "<val_data_dir>" }, { "name": "model_outdir", "type": "constant", "value": "<desired_outdir>" }, { "name": "learning_rate", "type": "float", "use_log_scale": true, "lower": 0.000001, "upper": 0.0001 }, { "name": "num_layers", "type": "int", "lower": 1, "upper": 9 }, { "name": "batch_size", "type": "ordered", "element_type": "int", "values": [16, 32, 64, 128, 256, 512], "sigma": 1 }, { "name": "warmup_type", "type": "ordered", "element_type": "string", "values": ["none", "linear", "quadratic", "exponential"], "sigma": 0.5 }, { "name": "optimizer", "type": "categorical", "element_type": "string", "values": [ "Adam", "SGD", "RMSprop" ] }, { "name": "epochs", "type": "constant", "value": 150 } ]
Make sure to set the hyperparameter space to what you desire, the above file is an example. The upper and lower describe the bounds of the hyperparameter. Hyperparameters of float, int, ordered, categorical, and constant types are supported, with ordered and categorical hyperparameters supporting float, int, and string types. Log scale exploration is also supported for float and int hyperparameter types. More about additional customization and methods can be found in the HPO Parameters Guide.
Supervisor setup
Supervisor is built on the Swift/T workflow language and the EMEWS Framework.
Supervisor is pre-installed on many relevant HPC systems. In these cases, you simply have to select the previously-developed “site” configuration already available.
If not already installed, follow these steps:
# Create environment
conda create --name IMPROVE python=3.9.16
conda activate IMPROVE
# Supervisor for running HPO/GA
git clone https://github.com/ECP-CANDLE/Supervisor.git
git checkout develop
cd Supervisor && PATH=$PATH:$(pwd)/bin
# swift-t
conda install --yes -c conda-forge -c swift-t swift-t
# python libraries
pip install numpy deap
Note that on clusters with specific MPI implementations, you must build Swift/T: https://swift-lang.github.io/swift-t/guide.html#_installation
Example
First, go into the directory where you have your configuration files:
cd ~/Experiment
Then, run the command:
supervisor ${location} ${workflow} ${config}
Running an HPO experiment on lambda. The model image is in /software/improve/images/. We will execute the command above with location set to conda and workflow set to GA. This will use the defaults from your conda environment.
supervisor conda GA cfg-1.sh
Debugging
While/after running HPO, there will be model.log files which contain the important information regarding that model’s run. They can be found at <candle_data_dir>/<model_name>/Output/EXP<number>/run_<number>. To debug, use a grep -r "ABORT" in the experiment directory <candle_data_dir>/<model_name>/Output/EXP<number> to find which run file which is causing the error in your workflow, cd run_<number> to navigate there, and cat model.log to observe the abort and what error caused it. Observing the MODEL_CMD (which tells the hyperparameters) and the IMPROVE_RESULT (which tells the evaluation of those hyperparameters) can also be helpful.
Results
After running HPO, there will be the turbine output and experiment directories. The turbine_output directory is found in the same directory as the config files and contains a final_result_<number> file which puts the HPO results in a table. The experiment directory is found at <candle_data_dir>/<model_name>/Output/EXP<number> and contains the output.csv file which has ALL the hpo parameters and results automatically parsed. The experiment directory also contains the hyperparams.json file you used to help remember the hyperparameter space you explored.
Analysis
To analyze the HPO run, there are two recommended methods. The first provides a ranking of hyperparameter choices. The second provides a ranking and visualization:
Firstly, the user could run the following commands in the experiment directory. The user is required to define the number of hyperparameters. In the example hyperparams.json file given, this would be 3 (learning_rate, batch_size, epochs). The sorted, unique choices of hyperparameters are put into a new
sorted_unique_output.csvfile.
num_hyperparams=3
num_columns=$((num_hyperparams + 1))
(head -n 1 output.csv && tail -n +2 output.csv | sort -t, -k$num_columns -n | uniq) > sorted_unique_output.csv
Secondly, the user could secure copy the output.csv file, then use google colab to show tables and plot. The secure copy command should be run in your terminal (not logged into Argonne’s computation system) as the following:
scp <user>@<computation_address>:~/path/to/your/output.csv \path\on\local\computer. For example, as secure copy command could look like:scp <username>r@polaris.alcf.anl.gov:~/data_dir/DeepTTC-testing/Output/finished_EXP060/output.csv \Users\<username>\Argonne\HPO. Note that this assumes the user is using Unix. If running a Unix-like system on Windows, the command will look likescp <user>@<computation_address>:~/path/to/your/output.csv /c/Users/username/Path/On/Local/Computer.
Once the file is secure copied to your local computer, it can be loaded into and used in google colab. For an example, follow the example and instructions here: https://colab.research.google.com/drive/1Us5S9Ty7qGtibT5TcwM9rTE7EIA9V33t?usp=sharing
High Level Framework
The Genetic Algorithm (GA) operates on principles derived from evolutionary biology to optimize model hyperparameters. In each generation of the GA, a new set of offspring is created by altering the current population’s hyperparameters, which are akin to biological genes. This alteration occurs through mutation, where random tweaks are made to individual’s hyperparameters, and crossover (or mating), where hyperparameters from two parents are mixed. In the case of HPO, each hyperparameter is analogous to a gene, and each set up hyperparameters an individual. Then, each offspring is evaluated for effectiveness using the IMPROVE_RESULT as a measure of fitness. Finally, there is a selection process that selects out high-performing individuals to carry on to the next generation. Over numerous generations, the population evolves, ideally converging towards an optimal set of hyperparameters.
In order to carry out a minimal-effort hyperparameter search using the defaults, only num_iterations and population_size need to be specified.
Parameters
The following is an explain of the parameters used for the genetic algorithm (GA), and how they interact with the hyperparameter space given. The GA workflow uses the Python DEAP package (http://deap.readthedocs.io/en/master) to optimize hyperparameters using a genetic algorithm.
The relevant parameters for the GA algorithm are defined in cfg-*.sh scripts (usually in cfg-my-settings.sh). These are:
NUM_ITERATIONS: The number of iterations the GA should perform.
POPULATION_SIZE: The maximum number of hyperparameter sets to evaluate in each iteration.
GA_STRATEGY: The algorithm used by the GA. Can be one of “simple” or “mu_plus_lambda”.
OFFSPRING_PROPORTION: The offspring population size as a proportion of the population size (this is rounded) (specifically for the mu_plus_lambda strategy)
MUT_PROB: Probability an individual is mutated.
CX_PROB: Probability that mating happens in a selected pair.
MUT_INDPB: Probability for each gene to be mutated in a mutated individual.
CX_INDPB: Probability for each gene to be swapped in a selected pair.
TOURNSIZE: Size of tournaments in selection process.
Note that these parameters all have defaults. For a minimal effort HPO run, only num_iterations and population_size need to be specified in order to define the size of the run.
Mechanics
The Genetic Algorithm is made to model evolution and natural selection by applying crossover (mating), mutation, and selection to a population in many iterations (generations).
In the “simple” strategy, offspring are created with crossover AND mutation, and the selection for the next population happens from ONLY the offspring. In the “mu_plus_lambda” strategy, offspring are created with crossover OR mutation, and the selection for the next population happens from BOTH the offpsring and parent generation. Also in the mu_plus_lambda strategy, the number of offspring in each generation is a chosen parameter, which can be controlled by the user through offspring_prop.
Mutation intakes two parameters: mut_prob and mut_indpb. The parameter mut_prob represents the probability that an individual will be mutated. Then, once an individual is selected as mutated, mut_indpb is the probability that each gene is mutated. For example, if an individual is represented by the array [11.4, 7.6, 8.1] where mut_prob=1 and mut_indpb=0.5, there’s a 50 percent chance that 11.4 will be mutated, a 50 percent chance that 7.6 will be mutated, and a 50 percent chance that 8.1 will be mutated. Also, if either of mut_prob or mut_indpb equal 0, no mutations will happen. The type of mutation we apply depends on the data type because we want to preserve data type under mutation and ‘closeness’ may or may not represent similarity. For example, gaussian mutation is rounded for integers to preserve their data type, and mutation is a random draw for categorical variables because being close in a list doesn’t equate to similarity.
Crossover intake two parameters: cx_prob and cx_indpb, which operate much in the same way as cx_prob and cx_indpb. For example, given two individuals represented by the arrays [1, 2, 3] and [4, 5, 6] where cx_prob=1 and cx_indpb=0.5, there’s a 50% chance that 1 and 4 will be ‘crossed’, a 50% chance that 2 and 5 will be ‘crossed’, and a 50% chance that 3 and 6 will be ‘crossed’. Also, if either mut_prov or mut_indpb equal 0, no crossover will happen. The definition of ‘crossed’ depends on the crossover function, which must be chosen carefully to protect data types. We use cx_Uniform, which swaps values such that [4, 2, 3], [1, 5, 6] is a possible result from crossing the previously defined individuals. One example of a crossover function which doesn’t preserve data types would be cx_Blend, which averages values.
Selection has various customizations, with tournaments being our implementation. In tournament selection, ‘tournsize’ individuals are chosen, and the individual with the best fitness score is selected. This repeats until the desired number of individuals are selected. Note that choosing individuals is done with replacement, which introduces some randomness to who is selected. Although unlikely, it’s possible for one individual to be the entire next population. It’s also possible for the best individual to not be selected as long as tournsize is smaller than the population. However, it is guaranteed that the worst ‘tournsize-1’ individuals are not selected for the next generation. Tournsize can be thought of as the selection pressure on the population.
Notes:
In the mu_plus_lambda strategy, cx_prob+mut_prob must be less than or equal to 1. This stems from how mutation OR crossover is applied in mu_plus_lambda, as opposed to mutation AND crossover in the simple strategy.
GPUs can often sit waiting in most implementations of the Genetic Algorithm because the number of evaluations in each generation is usually variable. However, with a certain configuration, the number of evaluations per generation can be kept at a constant number of your choosing. By using mu_plus_lambda, the size of the offspring population is made through the chosen parameter of offspring_prop. Then, by choosing cx_prob and mut_prob such that cx_prob+mut_prob=1, every offspring is identified as a ‘crossed’ or mutated individual and evaluated. Hence, the number of evaluations in each generation equals lambda. Note that because of cx_indpb and mut_indpb, an individual may be evaluated with actually having different hyperparameters. This also means that by adjusting mut_indpb and cx_indpb, the level of mutation and crossover can be kept low despite cx_prob+mut_prob being high (if desired). Note that the number of evaluations per generation can be kept constant in the simple strategy as well, but the number of evals has to be the population size.
The default values are: NUM_ITERATIONS=5 | POPULATION_SIZE=16 | GA_STRATEGY=mu_plus_lambda | OFFSPRING_PROP=0.5 | MUT_PROB=0.8 | CX_PROB=0.2 | MUT_INDPB=0.5 | CX_INDPB=0.5 | TOURNSIZE=4
See https://deap.readthedocs.io/en/master/api/algo.html?highlight=eaSimple#module-deap.algorithms for more information.